Abstract
Introduction Clonal hematopoiesis of indeterminate potential (CHIP) also known as age-related clonal hematopoiesis (ARCH) is a myeloid neoplasm composed of clonal bone marrow stem cells in non-cytopenic patients that is present in >10% of people over the age of 70. Most studies of CHIP/ARCH to date have relied on sequencing of peripheral blood as a surrogate for less easily obtainable bone marrow, however, it is unknown how well blood represents the full clonal heterogeneity of bone marrow, especially across multiple bone marrow sites. Further, high-sensitivity sequencing studies of the blood have shown that low-level clonal hematopoiesis can be detected in nearly all older individuals. In this study we sought to characterize regional clonal heterogeneity in the bone marrow and blood by sequencing marrow collected from multiple sites in older autopsy patients.
Methods We identified a prospective series of 10 recently deceased, non-cytopenic patients over the age of 65 (median age 75, range 66-93) who were consented for hospital autopsy and had no history of cancer or prior chemotherapy. Causes of death included myocardial infarction (5 patients) and ruptured aortic aneurysm (2 patients). Bone marrow was collected from 10 different sites on each patient including bilateral iliac crests, bilateral ribs, bilateral femurs, bilateral clavicles, sternum, and thoracic vertebra; central blood was collected from the heart and normal control tissue was collected from the brain frontal cortex. DNA was extracted from blood and marrow and subjected to both exome sequencing and high-coverage (~50,000x), error-corrected sequencing using a panel of 47 recurrently mutated genes (MyeloSeqHD) with an estimated sensitivity of 0.25% VAF. All sequencing data was analyzed using the Illumina Dragen platform.
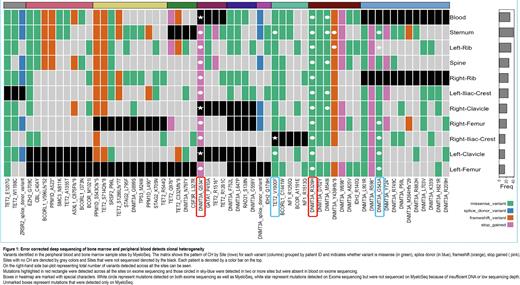
Results CHIP/ARCH was detected in the blood of 2 out of 10 patients by exome sequencing. Both patients had DNMT3A mutations (p.Q534* and p.R326H) with VAFs of 24% and 19%, respectively. These mutations were detected across all 10 of the sampled bone marrow sites and were not detected in germline control tissue. We next evaluated the bone marrow to determine whether CHIP/ARCH was more readily detectable from marrow compared to blood using exome sequencing and identified an additional two patients with mutations in DNMT3A, and TET2 present in 2 or more bone marrow sites but not blood. Exome data showed no evidence of chromosomal gains or losses in the blood or any bone marrow site. To identify low level clonal mutations, we then used high sensitivity targeted sequencing (MyeloSeqHD) on the same set of samples and detected 54 additional clonal hematopoiesis variants across all 10 patients (mean 2.3 variants per site per patient; median VAF=0.39%). Mutations were present predominantly in DNMT3A, TET2, PPM1D, and ASXL1 genes. (Fig.1). The maximum number of detected variants across the patients were in the sternum followed by left-rib, blood, spine, right-rib, left-iliac-crest, right-clavicle, right-femur, right-iliac-crest, left-clavicle, and left-femur respectively. 70% of the variants detected at marrow sites were present in the blood.
Conclusion To our knowledge, this is the first study to evaluate clonal hematopoiesis across multiple bone marrow sites and blood. Using high sensitivity error-corrected sequencing we demonstrate that low level CHIP/ARCH can be detected in all patients included in this study and show that these clones can be detected across multiple bone marrow sites and blood, providing further evidence that these low-level mutations, which have similarly been reported by other groups, represent bona fide clonal hematopoiesis. We also show that the blood and marrow contain many small clonal populations and that clonal diversity is greatest in the sternum and least in the distal skeleton. Finally, we show that at least 70% of low-level clonal hematopoiesis detected across multiple bone marrow sites can be detected in the blood, consistent with the idea that the marrow and blood are generally homogenous, and that blood is an adequate surrogate for bone marrow for the majority of variants.
Disclosures
Spencer:Wugen Inc.: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal